We are delighted to announce that members of our Microbiome Dynamics Group within the Cluster of Excellence “Balance of the Microverse” have contributed to deciphering a mechanism that determines how the gut microbiome processes health-promoting plant compounds. This work provides an important foundation for fostering holobiome balance through targeted, individual interventions.
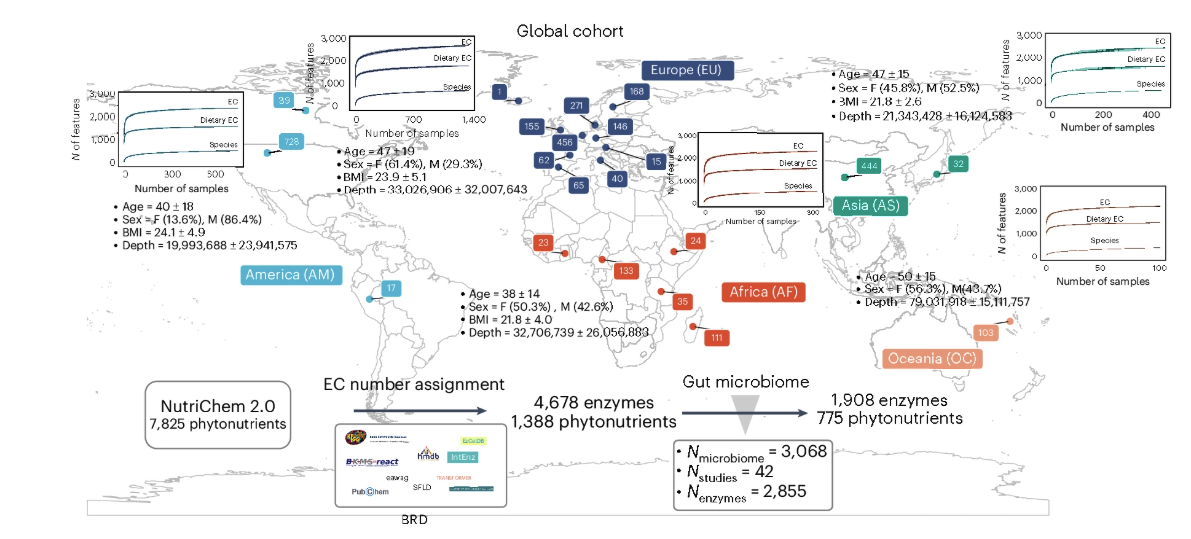
Our latest publication, “Gut microbiome-mediated transformation of dietary phytonutrients is associated with health outcomes”, presents a comprehensive, multi-database analysis linking gut microbial enzymes to the transformation of 775 phytonutrients from edible plants across 3,068 global public human microbiomes.
In vitro validation highlights specific gut bacterial species, including Eubacterium ramulus, and reveals substantial interpersonal and geographical variability in phytonutrient biotransformation capacity. Furthermore, machine-learning analyses conducted on 2,486 public case–control microbiomes demonstrate that enzyme abundances involved in phytonutrient modification can distinguish health states across multiple disease contexts, underscoring the potential for personalized, diet-guided interventions.
By integrating diverse datasets on enzymatic reactions and health benefits with large-scale metagenomic data, we show that microbiome-encoded biotransformation potential is closely associated with health phenotypes and disease contexts. Using metagenomic and metatranscriptomic analyses in germ-free and specific-pathogen-free (SPF) mouse models, we further validate the link between microbial enzymes and the anti-inflammatory activity of common edible plants.
Key Findings
- Global scope of phytonutrient biotransformation: Transformation of 775 plant phytonutrients is associated with enzymes encoded by diverse gut microbes across thousands of samples.
- Species-level validation: In vitro assays confirm biotransformation activity by gut bacteria, notably Eubacterium ramulus.
- Variability across individuals and regions: Biotransformation potential shows high interpersonal and geographical variability.
- Health-state discrimination via enzymes: Machine-learning models using microbial enzyme abundances related to phytonutrient modification distinguish health status in multiple disease contexts.
- In vivo validation of anti-inflammatory activity: Metagenomics and metatranscriptomics in SPF and germ-free mice support the association between microbiome-encoded enzymes and anti-inflammatory effects of edible plants.
Methods at a Glance
- We integrated multiple databases containing enzymatic reactions and health-benefit information.
- We analyzed 3,068 global public human microbiomes to map phytonutrient biotransformation.
- We performed in vitro assays to validate activity of gut species (e.g., Eubacterium ramulus).
- We built machine-learning models on 2,486 public case–control microbiomes to relate enzyme abundances to health status.
- We used metagenomics and metatranscriptomics in specific-pathogen-free and germ-free mouse models to validate functional associations.
Significance for Health, Environment, and Society
These findings suggest that individual and regional dietary responses are partly shaped by microbiome-encoded biotransformation capabilities. This opens a path toward precision nutrition strategies that tailor plant-rich diets to a person’s microbial potential, with implications for disease prevention, therapeutic monitoring, and sustainable dietary guidelines.
About the Publication
MBD members Dr. Bastian Seelbinder, Dr. Ana Depetris Chauvin, Prof. Gianni Panagiotou, Yueqiong (Bernard) Ni (alumni) and Lu Zhang (alumni) and external collaborators contributed to the study. We are very grateful to all our collaborators and funding sources from the German agency DFG who make this work possible.
Journal: Nature Microbiology (Impact factor 19.4)
Publication Date: 3rd of December, 2025
DOI: 10.1038/s41564-025-02197-z
Open Access/Access Options:
CC BY-NC-ND 4.0
Acknowledgments
This work was supported by the DFG - Deutsche Forschungsgemeinschaft under the project “MICROVERSE” (EXC 2051 – Project-ID 390713860).
Original Publication
Zhang L#, Marfil-Sánchez A#, Kuo T-H, Seelbinder B, van Dam L, Depetris-Chauvin A, Jahn L J, Sommer M O A, Zimmermann M, Ni Y*,
Panagiotou G* (2025). Gut microbiome-mediated transformation of dietary phytonutrients is associated with health outcomes.
Nature Microbiology. DOI: 10.1038/s41564-025-02197-z
# These authors contributed equally to this work.
* Corresponding authors